



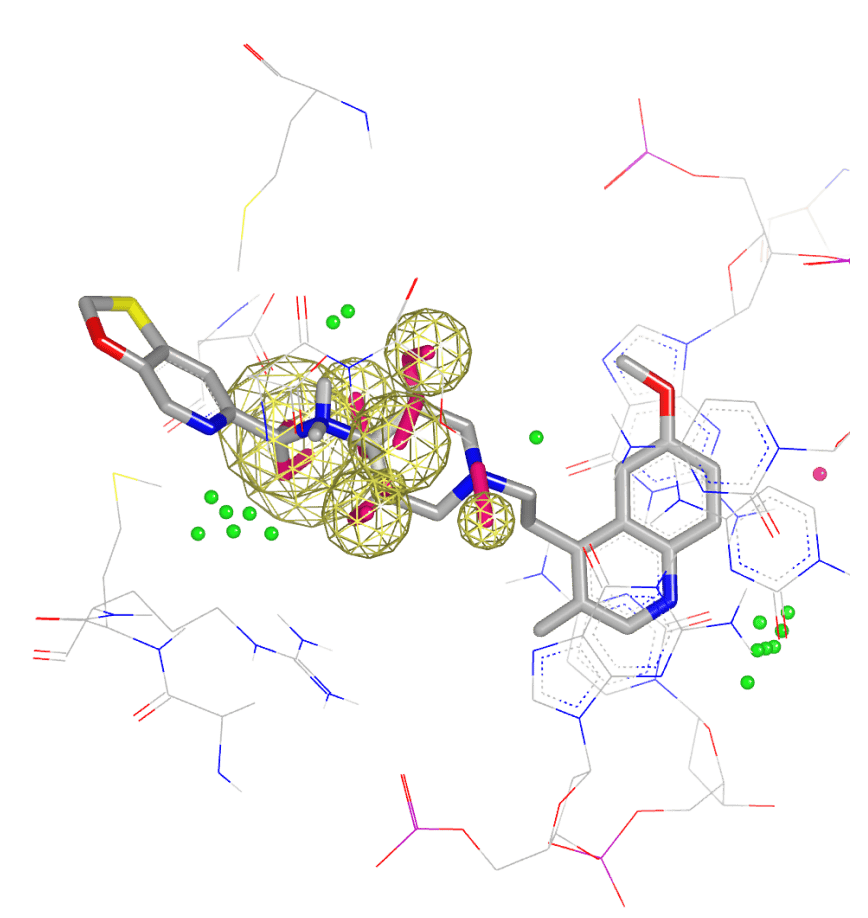
SCIENCE
Use physics-based design to advance
- Biomolecular target exploration
- Hit identification, hit-to-lead, and lead optimization
- Free energy predictions
- Pharmaceutical formulations
SPEED
Break the speed barrier searching billions of available molecules
- Ligand- and structure-based virtual screening
- Molecular dynamics (MD) simulations
- Affinity predictions
- Quantum chemistry calculations
2D similarity search on
billions of ligands in
Seconds
3D similarity search on
billions of ligands in
Minutes
Dock billions of
ligands in
Hours
SCALE
Accelerate science to the speed of now
- Screen ultra-large scale compound databases
- Perform long timescale MD simulations on large systems
- Improve impact using rigorous theory & efficient algorithms
- Design & screen antibody libraries using NGS
RESOURCES
Glimpse the Future through News, Events, Webinars and more


