
VISUALIZATION & COMMUNICATION OF MODELING RESULTS
VIDA
Molecular modeling is more than just generating numbers. It is about understanding, rationalizing, predicting, and, most importantly, communicating results. Ultimately, modeling efforts are pointless if they don't influence the target audience. To maximize impact, the audience itself needs an easy-to-use tool to study the appropriate data without overwhelming detail. VIDA offers this solution.
For more detailed information on VIDA, check out the link below:
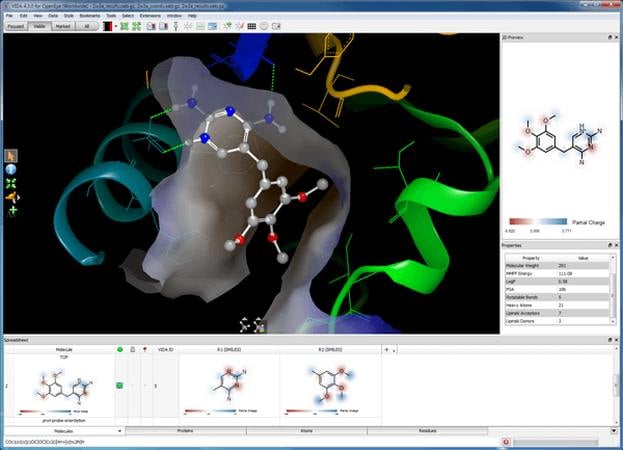
Visualizing docking results with VIDA's Grapheme™-enabled spreadsheet.
Features
- Advanced 3D graphics for high quality molecular visualizations
- Robust handling and easy browsing of large molecular data sets
- Chemically aware spreadsheet featuring real-time interaction with 1D (SMILES), 2D, and 3D displays
- Preset, customizable views for docking (FRED) and ligand-based similarity searches (ROCS®, EON)
- Scriptable with Python enabling easy customization and extensibility
- All OpenEye Toolkits can be accessed directly through Python scripting interface
- Telemodeling option that allows for interactive sharing of sessions between multiple users over a network
- PDF export of molecular spreadsheet information for easy off-line analysis and discussion
- Supported on Linux, Windows® and Mac OS
ROCS X: AI-Enabled Molecular Search Unlocks Trillions
Webinar: OpenEye's Free energy prediction for drug discovery: Ideas at breakfast, discoveries by lunch
Science Brief: Binding Free Energy Redefined: Accurate, Fast, Affordable
CUP XXV - Santa Fe March 10-12, 2026
Webinar: Novel Hits from Beyond the Known: ROCS X
Webinar: OpenEye's Free energy prediction for drug discovery: Ideas at breakfast, discoveries by lunch
Science Brief: Binding Free Energy Redefined: Accurate, Fast, Affordable
CUP XXV - Santa Fe March 10-12, 2026
Webinar: Novel Hits from Beyond the Known: ROCS X
Resources
Glimpse the Future through News, Events, Webinars and more
News
ROCS X: AI-Enabled Molecular Search Unlocks Trillions
BOSTON, OpenEye miniCUP—Sept. 25, 2025—Cadence Molecular Sciences (OpenEye), a business unit of Cadence (Nasdaq: CDNS), announced today at miniCUP Boston, the launch of ROCS X, an AI-enabled virtual screening solution that allows scientists to conduct 3D searches of trillions of drug-like molecules.
Read now
Webinar
Webinar: OpenEye's Free energy prediction for drug discovery: Ideas at breakfast, discoveries by lunch
Webinar: OpenEye's Free energy prediction for drug discovery: Ideas at breakfast, discoveries by lunch We are continuing our 2025 miniWEBINAR series with our November session led by Chris Neale, Ph.D., who leads the molecular binding affinity solution group at OpenEye, Cadence Molecular Sciences. His expertise and interests include method development for structure-based drug design, cellular signal transduction, and uncertainty quantification. Chris is also an editorial board member at the Biophysical Journal.
Read now


