
SZMAP/GamePlan
All protein-ligand binding interactions occur in an aqueous environment, therefore predicting the effects of solvent molecules within the binding site is a necessary step in understanding binding affinity. However, the interactions of protein and ligand together with water molecules can have complex and non-obvious effects, which can complicate and obfuscate a lead-optimization campaign.
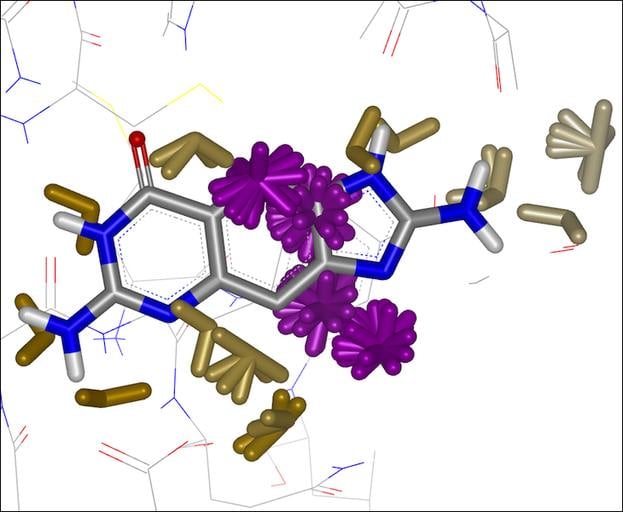
SZMAP is OpenEye’s application to help modelers and medicinal chemists understand the role of water in molecular interactions such as ligand binding. Using water as a lens, SZMAP can provide the user with insight on critical features of a binding site, predict where and how neighboring waters can influence binding of a ligand, or generate ligand modification hypotheses designed to better exploit specific regions on a binding site. The semi-continuum [1, 2] approach used in SZMAP combines one explicit water molecule with robust continuum solvent methods [3], classical statistical mechanics, and thorough sampling plus the ability to analyze specific water orientations.
Features
- Maps various thermodynamic properties across holo, apo and ligand structures
- Identifies key water sites and their orientational preferences
- Predicts changes in water activity on ligand binding
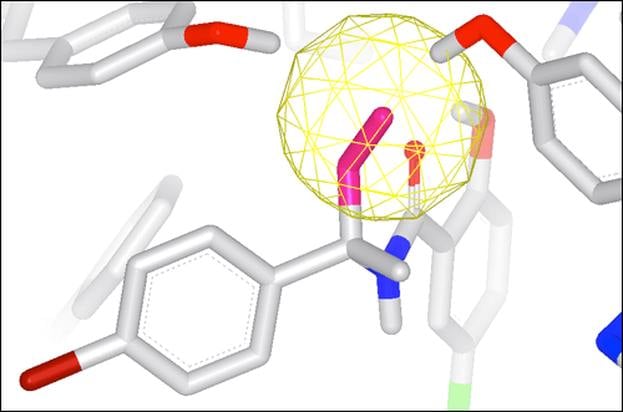
- Performs very rapid calculations at specified sets of coordinates such as atom centers of bound ligands
- GamePlan allows for rapid calculation and analysis of SZMAP result to suggest ligand modification hypotheses
- Includes a facility for writing data to multiple grid formats and performing mathematical operations on SZMAP grids
- Results can be visualized in 3D using VIDA
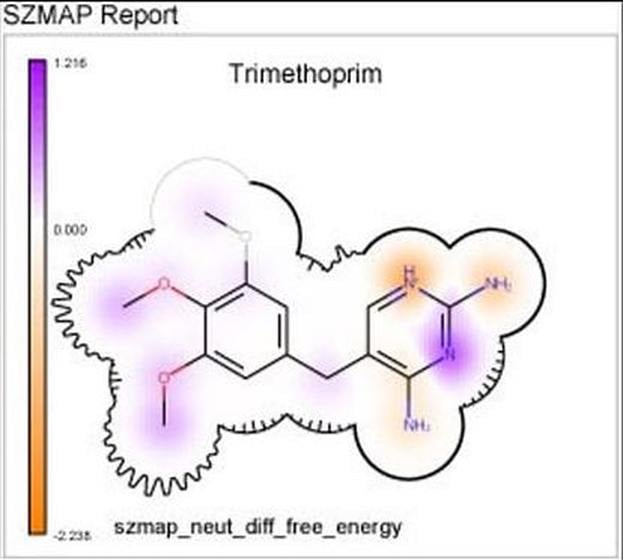
- 2D Grapheme™ representations present SZMAP results in a format that is natural for a chemist
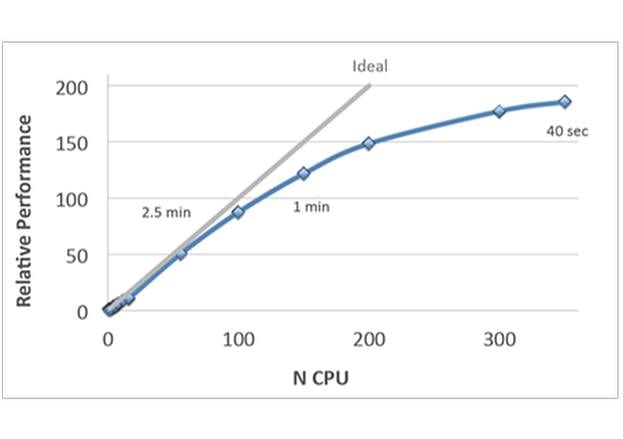
- Improved speed and full parallelization through MPI
GamePlan
Integrated into SZMAP is GamePlan, a unique approach to ligand modification/optimization based on the energetics of water at points in a protein binding site close to the ligand of interest. GamePlan provides easy to interpret suggestions for manipulations of a molecular scaffold or additions/extensions to that scaffold to improve binding affinity using an understanding of both the enthalpic and the entropic contributions to water stability.
For more detailed information on SZMAP, check out the link below:
DocumentationLearn more
OpenEye's SZMAP/GamePlan can suggest ligand modifications to optimize binding interactions, thus speeding up your drug-design process.
Watch the on-demand webinar on SZMAP & GamePlan: Optimize your lead molecules by understanding water
References
- Calculation of the thermodynamic properties of aqueous electrolytes to 1000.degree.C and 5000 bar from a semicontinuum model for ion hydration Tanger, J.C., Pitzer, K.S., J. Phys. Chem. 1989, 93, 4941-4951.
- Continuum based calculations of hydration entropies and the hydrophobic effect Rashin, A.A., Bukatin, M.A., J. Phys. Chem. 1991, 95, 2942-2944.
- A Smooth Permittivity Function for Poisson-Boltzmann Solvation Methods Grant, J.A., Pickup, B., Nicholls, A., J. Comp. Chem. 2001, 22, 608-640.
Webinar: OpenEye's Free energy prediction for drug discovery: Ideas at breakfast, discoveries by lunch
Science Brief: Binding Free Energy Redefined: Accurate, Fast, Affordable
CUP XXV - Santa Fe March 10-12, 2026
Webinar: Novel Hits from Beyond the Known: ROCS X
Resources
Glimpse the Future through News, Events, Webinars and more
News
ROCS X: AI-Enabled Molecular Search Unlocks Trillions
Webinar
Webinar: OpenEye's Free energy prediction for drug discovery: Ideas at breakfast, discoveries by lunch


