The number of drug-like molecules that could exist is staggering. Estimates range as high as 1060, more than the number of atoms in the Universe.
Commercially available collections of synthetically accessible drug-like molecules now number in the billions, and these collections are expanding regularly, while virtual collections of compounds can exceed a trillion molecules.
In order to address the challenges posed by the rapid increase in the size of accessible chemical space, OpenEye Scientific has been focused on accelerating our molecular search algorithms for both the CPU and GPU, specifically similarity search in GraphSim TK and substructure search in OEChem TK. As a result of these recent upgrades, you now are able to search a billion molecules in seconds.
Similarity Search
Generating Fingerprints
Fingerprinting provides an elementary encoding of molecular structures. Even though fingerprints can only represent local structural features and not their relative positions inside molecules, they have been proven to be very successful in a range of similarity and diversity studies.
GraphSim TK provides three fingerprint types that exhaustively encodes molecules: path (Daylight-like), circular (ECFP-like), and tree. These fingerprints can be pre-generated in a multi-threaded process and stored in a fingerprint binary file.
Searching Fingerprints
The process of the fast fingerprint search is illustrated in Figure 1. There are three ways to search the pre-generated fingerprints: in-memory, memory-mapped, and CUDA.
The in-memory (CPU) method involves pre-loading all fingerprints into memory and performing the search in-memory. This presents the fastest CPU search at the expense of load time and limitation of the search by the size of the memory.
When using the memory-mapped (CPU) method there is no load time penalty or memory limitation but the search itself is marginally slower.
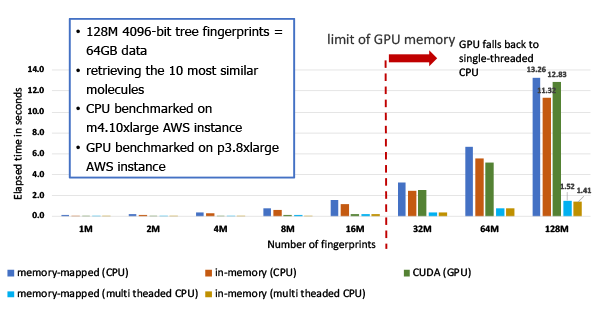
These two search modes can now be performed utilizing multiple threads (via the new SortedSearch method). See the performance improvement shown in Figure 2.
The new method not only returns the best similarity matches but also keeps the score distribution for the whole dataset and provides access to the search progress.
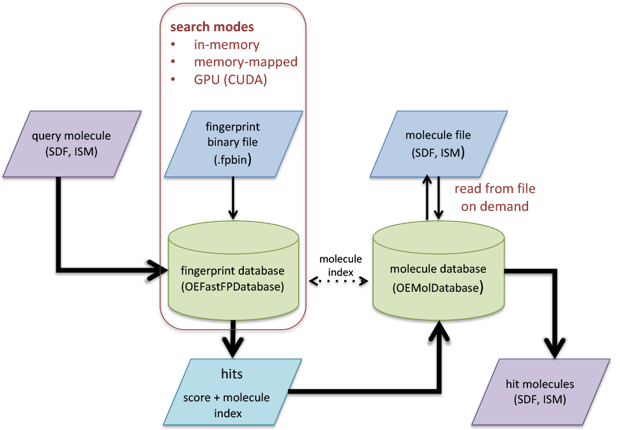
Figure 1: Schematic representation of fast fingerprint search process.
The CUDA mode is a GPU-enabled calculation that provides 200x faster calculation than the single-threaded CPU modes. CUDA mode involves pre-loading all fingerprints into GPU memory prior to performing similarity calculations. The CUDA mode is suitable for N x N similarity calculation but searches are limited by GPU memory availability and will fall back to the memory-mapped CPU mode if the entire set of fingerprints cannot be preloaded into the GPU memory.
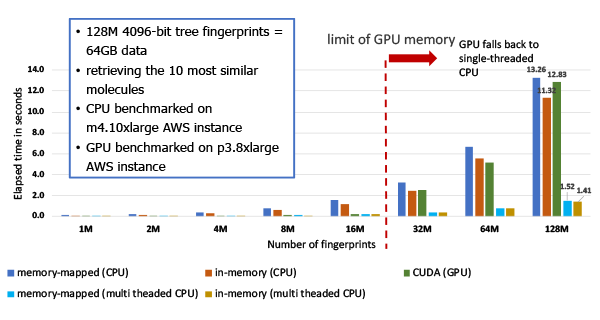
Figure 2: The performance of searching 4096 bit tree fingerprints and retrieving top 10 hits.
GraphSim TK provides a straightforward and user friendly API (see Figure 3) to perform similarity searches. Functionality to perform N x N searches or to build sparse matrices for clustering is also available.

Figure 3: Python code snippet of fast fingerprint search process.
Substructure Search
Substructure search methods identify compounds that contain a user-defined partial structure (i.e. pattern of bonds and atoms) irrespective of the environment in which the query occurs. In graph theory, this problem is called subgraph isomorphism, a problem class that computational theorists call NP-complete.
In OEChem TK several different approaches are implemented to accelerate the substructure search algorithm:
- Optimizing expressions of query molecules
- Pre-screening
- Multi-threaded search
Pre-Screening
Screens are bit vectors that encode both global and local features of the query and the target molecules. This is one of the most effective ways to accelerate the search since target molecules that clearly cannot be matched to the query can be rapidly eliminated. Only molecules that pass the screening phase are subjected to the more expensive atom-by-atom validation.
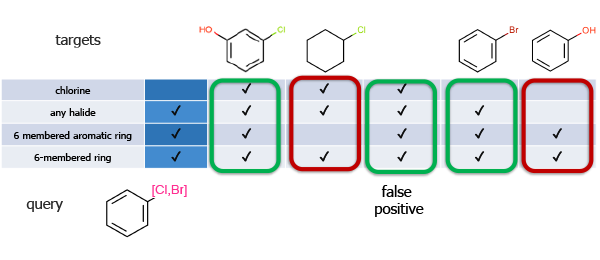
Figure 4: If a query molecule contains a feature (bit) that is not present in a target molecule, then the target can be eliminated from further consideration.
Screening paradox: larger molecules take longer to search (atom-by-atom), however they have more unique features and, hence, are easier to screen out.
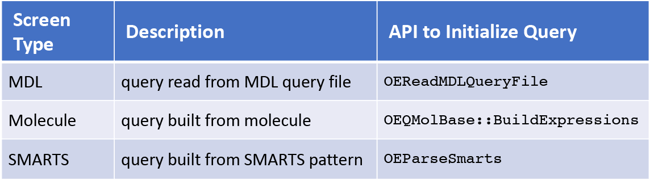
Figure 5: OEChem TK provides three types of built-in screens based on the source of the query molecule.
All screen types are rigorously tested using diverse sets of queries and multiple molecule databases to increase their efficiency (see Figure 5) and ensure that they do not eliminate true positive matches.
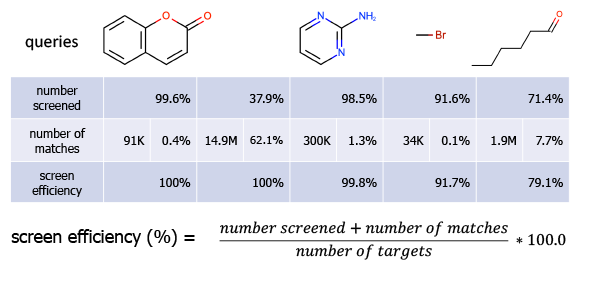
Figure 6: Screen efficiency. 100.0% means that no false positives were detected i.e. each molecule that passed the screening phase was a real match.
Substructure search databases are OEB binary files with prepared molecules and pre-built screens. The database generation process is multi-threaded enabling speed-ups that scale.
The substructure search engine can be initialized in two modes: in-memory and molecule-database.
The in-memory mode provides the fastest way to search a dataset, but it is memory-intensive as it holds both screens and molecules in memory.
The molecule-database mode keeps only the screens in memory; only unscreened molecules have to be loaded into the memory on demand. This method can be slower but uses significantly less memory, allowing users to search larger datasets.
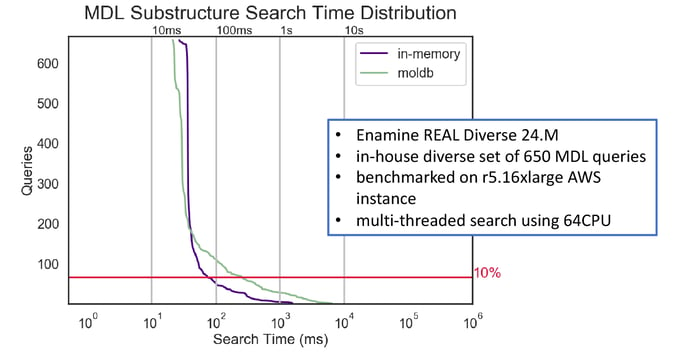
Figure 7: The performance of screen-based multi-threaded substructure search using MDL queries.
To learn more about GraphSim TK and OEChem TK, as well as our other programming libraries for creating custom applications, scripts and web services, please visit our Toolkit Development Platform page.
Provide a comment below on this blog post.