
FastROCS
GPU-accelerated Shape Similarity Search
Scale your virtual screening and lead hopping to billions and beyond with FastROCSTM.
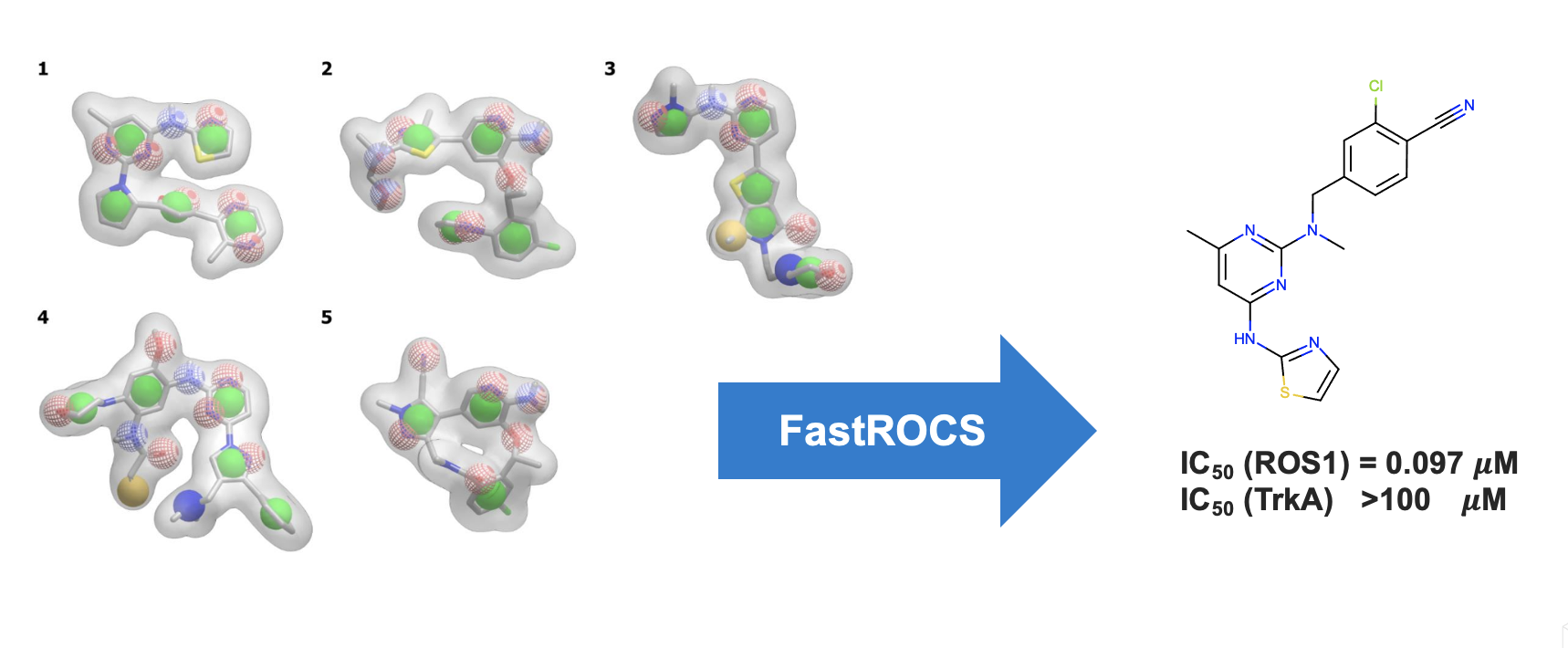
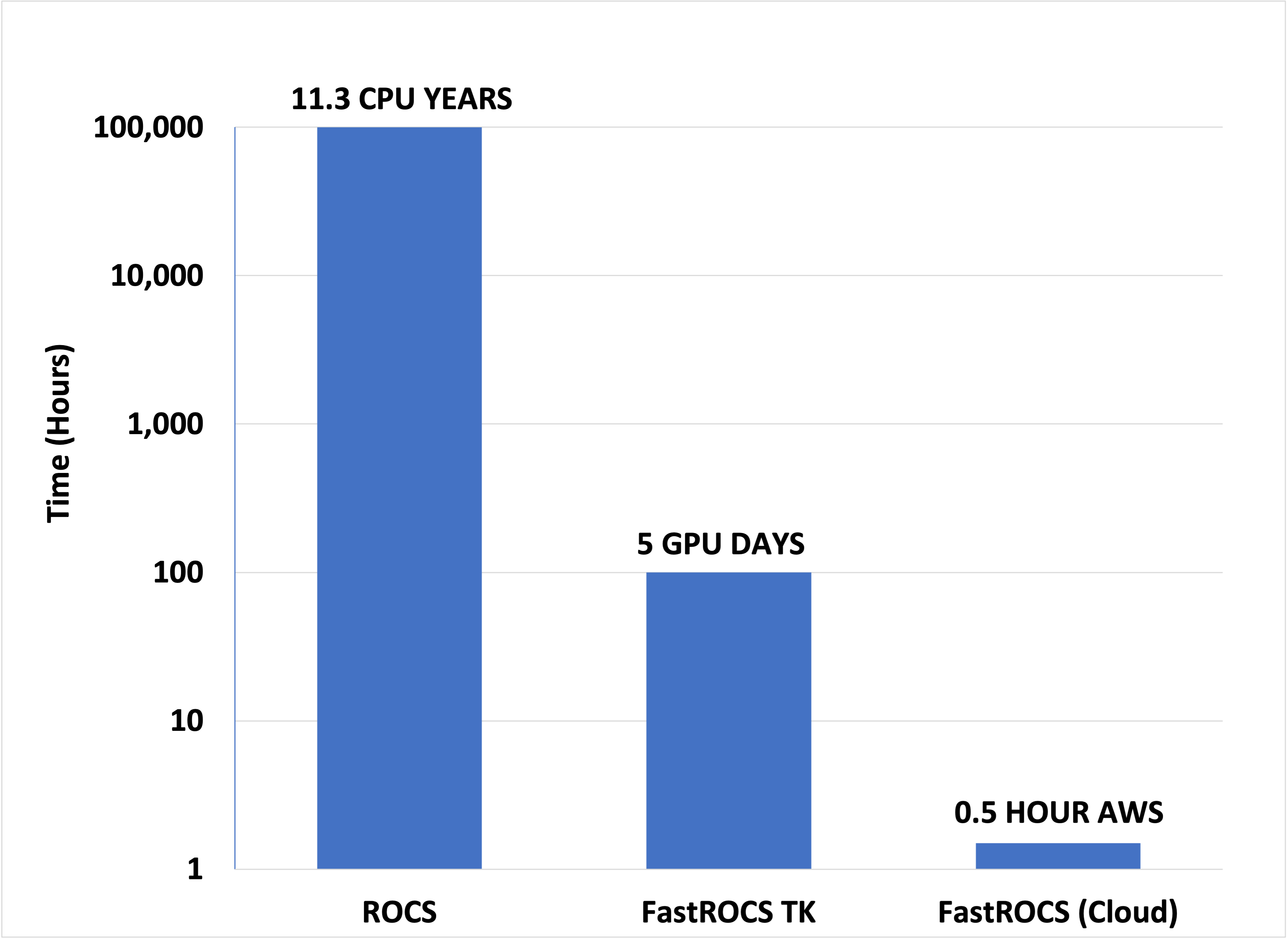
Get near-instantaneous results for your virtual screening and lead hopping needs. FastROCS performs 3D alignment and scoring, based on a robust Gaussian-based description of shape and chemical features, at speeds approaching those of 2D methods. This unparalleled speed allows you to conduct highly accurate 3D shape and chemical feature similarity calculations on millions of molecules within seconds, delivering hits that are highly structurally diverse compared to those from 2D searches.
Our prospective study describes how conceptually simple shape-matching approaches can identify potent and selective compounds by searching ultralarge virtual libraries, demonstrating the applicability of such workflows and their importance in early drug discovery. -- Petrovic et al. (JCIM, 2022)
Features
- Highly effective virtual screening based on robust and reliable science
- Get near-instantaneous results. Processes millions to hundreds of millions of conformations per second
- Launch jobs and view results using VIDA desktop visualizer or Orion web interface
- Overlay results are intuitive and visually informative
- Support both Gaussian and grid-based shape query
- FastROCS Plus. Seamlessly combine ligand- and structure-based approaches in one step
Seamlessly combine 3D ligand- and structure-based screening

FastROCS™ Plus offers a comprehensive, turnkey solution that integrates 3D ligand- and structure-based screening into a streamlined, automated workflow
FastROCS Plus provides scientists with a high-performance GPU-accelerated shape-based screening tool designed for rapid, large-scale molecular comparison. Built on the proven ROCS (Rapid Overlay of Chemical Structures) technology, FastROCS Plus leverages parallel GPU processing to deliver ultra-fast shape and chemical feature similarity calculations across millions of compounds in seconds. Ideal for virtual screening campaigns and early-stage drug discovery, it enables researchers to explore chemical space more efficiently, prioritize candidates faster, and make data-driven decisions with confidence. Combining speed, scalability, and precision, FastROCS Plus delivers better hits; because better hits start with better screening.
How Scientists are Using FastROCS
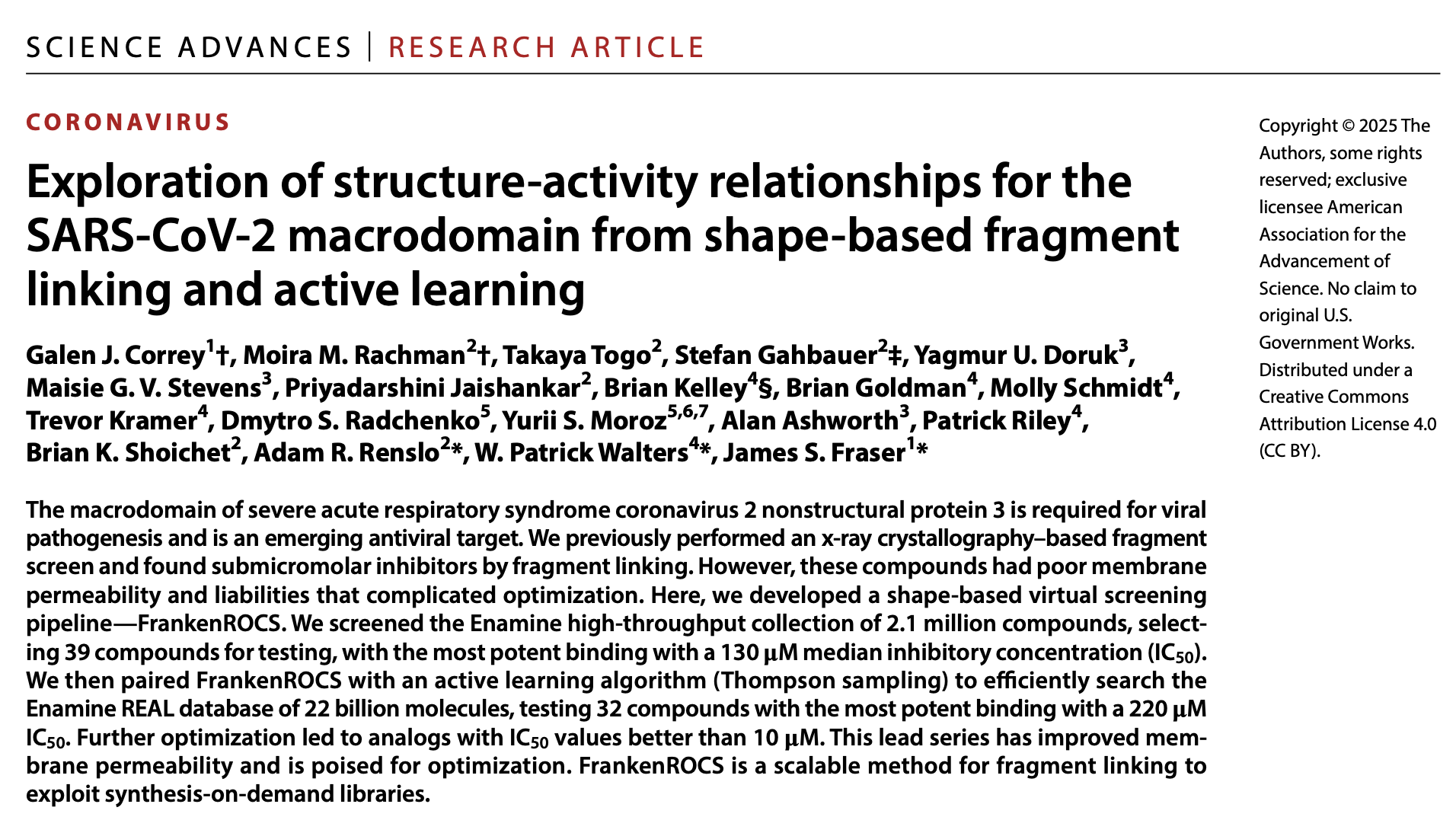
FrankenROCS, a novel shape-based virtual screening pipeline, was developed by teams at UCSF and Relay Therapeutics to address limitations in lead optimization, particularly for SARS-CoV-2 macrodomain inhibitors with poor membrane permeability. This pipeline integrates OpenEye's FastROCS for 3D similarity searching with an active learning algorithm (Thompson sampling).
This methodology enabled the efficient exploration of the 22-billion-molecule Enamine REAL database, overcoming the prohibitive scale of traditional screening. The approach successfully identified submicromolar inhibitors with improved cell permeability and metabolic stability, achieving significant potency gains through fragment linking. This demonstrates a scalable strategy for discovering structurally validated, drug-like candidates from synthesis-on-demand libraries. Link to publication.
FAQs
-
FastROCS can be accessed both in its toolkit form (FastROCS TK) and on OpenEye's cloud-based Orion Modeling Platform.
-
ROCS® is a comprehensive tool for fast shape and chemical feature similarity, able to process hundreds of molecules per CPU second.
FastROCS combines the state-of-the-art science from ROCS with the speed of the GPU. With FastROCS, you can search virtual libraries of billions of compounds or cluster millions of molecules by shape. -
FastROCS TK supports both Gaussian and grid-based shape queries, expanding the types of queries inputs to include active site negative images, composite queries combining grids and molecules, and queries derived from crystallographic density.
-
FastROCS Plus extends the search capabilities of FastROCS by using consensus scoring with high-speed docking. FastROCS Plus seamlessly combines ligand- and structure-based approaches in one step.
FastROCS and FastROCS Plus - trusted science, extreme speed, and unlimited scale! Find your next drug candidate, faster.
Modeling
The Modeling suite of toolkits provides the core functionality underlying OpenEye's defining principle that shape & electrostatics are the two fundamental descriptors determining intermolecular interactions. Many of the toolkits in the Modeling suite are directly associated with specific OpenEye applications and can, therefore, be used to create new or extend existing functionality associated with those applications.
- OEChem TK Core chemistry handling and representation as well as molecule file I/O
- FastROCS™ TK Real-time shape similarity for virtual screening, lead hopping & shape clustering
- OEDocking TK Molecular docking and scoring
- Omega TK Conformer generation
- Shape TK 3D shape description, optimization, and overlap
- SiteHopper TK Rapid comparison of protein binding sites
- Spicoli TK Surface generation, manipulation, and interrogation
- Spruce TK Protein preparation and modeling
- Szybki TK Force field based focused optimization and entropy estimations
- OEFF TK Force fields and general optimization tools
- Szmap TK Understanding water interactions in a binding site
- Zap TK Calculate Poisson-Boltzmann electrostatic potentials
- Bioisostere TK 3D fragment similarity, lead optimization, and analog generation
- Eon TK Molecular shape and electrostatic overlap
Cheminformatics
The Cheminformatics suite of toolkits provides the core foundation upon which all the OpenEye applications and remaining toolkits are built.
- OEChem TK Core chemistry handling and representation as well as molecule file I/O
- OEDepict TK 2D Molecule rendering and depiction
- Grapheme™ TK Advanced molecule rendering and report generation
- GraphSim TK 2D molecular similarity (e.g. fingerprints)
- Lexichem™ TK Name-to-structure, structure-to-name, foreign language translation
- MolProp TK Molecular property calculation and filtering
- Quacpac TK Tautomer enumeration and charge assignment
- OEMedChem TK Matched molecular pair analysis, fragmentation utilities, and molecular complexity metrics
- Saiph TK: Extracting, transforming, and loading (ETL) files and records
Learn More
OpenEye’s shape searching technology helps you use maximize 3D ligand information to speed up your drug-design process.
WATCH OpenEye’s scientist, Mark McGann, Ph.D., describes the methods behind FastROCS and FastROCS Plus.
References
- A fast method of molecular shape comparison: A simple application of a Gaussian description of molecular shape Grant, J.A., Gallardo, M.A., Pickup, B., J. Comp. Chem., 1996, 17, 1653.
- A shape-based 3-D scaffold hopping method and its application to a bacterial protein-protein interaction Rush, T.S., Grant, J.A., Mosyak, L., Nicholls, A., J. Med. Chem., 2005, 48, 1489.
- Comparison of Shape-Matching and Docking as Virtual Screening Tools Hawkins, P.C.D., Skillman, A.G., Nicholls, A., J. Med. Chem., 2007, 50, 74.
- Assessment of Scaffold Hopping Efficiency by Use of Molecular Interaction Fingerprints Venhorst, J., Nunez, S., Terpstra, J.W., Kruse, C.G., J. Med. Chem., 2008, 51, 3222.
- Multiple protein structures and multiple ligands: effects on the apparent goodness of virtual screening results Sheridan, R.P., McGaughey, G.B., Cornell, W.D., J. Comput. Aided Mol. Des., 2008, 22, 257.
- Lessons in Molecular Recognition. 2. Assessing and Improving Cross-Docking Accuracy Sutherland, J.J., Nandigam, R.K., Erickson, J.A., Vieth, M. J. Chem., Inf. Model, 2007, 49, 1715.
Webinar: OpenEye's Free energy prediction for drug discovery: Ideas at breakfast, discoveries by lunch
Science Brief: Binding Free Energy Redefined: Accurate, Fast, Affordable
CUP XXV - Santa Fe March 10-12, 2026
Webinar: Novel Hits from Beyond the Known: ROCS X
Resources
Glimpse the Future through News, Events, Webinars and more
News
ROCS X: AI-Enabled Molecular Search Unlocks Trillions
Webinar
Webinar: OpenEye's Free energy prediction for drug discovery: Ideas at breakfast, discoveries by lunch



