
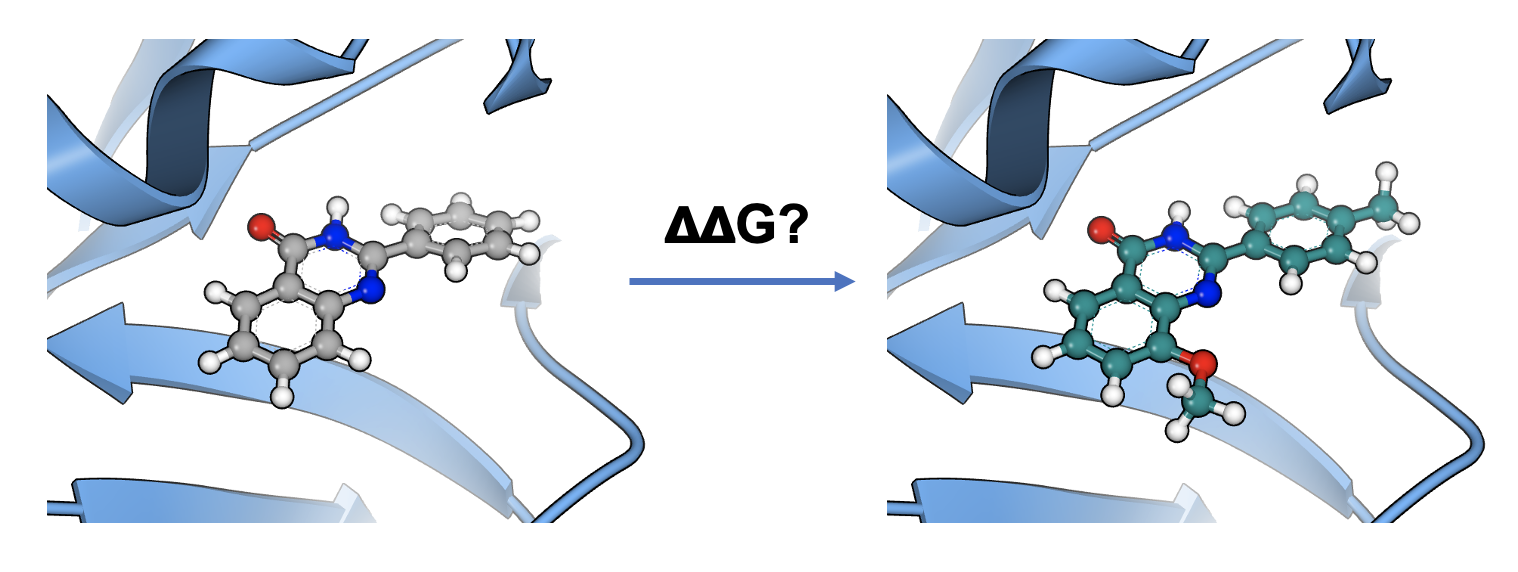
Binding Free Energy Predictions -Accurate, Fast, and Affordable
OpenEye provides efficient and effective solutions for estimating the binding free energies of protein–ligand complexes.
Free Energy Nonequilibrium Switching (FE-NES) calculates ligand–protein binding free energies using the non-equilibrium switching technique, advanced by Gapsys and De Groot (2020) from the Max Planck Institute. Scientific and technical enhancements in FE-NES make it higher throughput and more cost-effective than traditional equilibrium methods such as free energy perturbation (FEP), which require extensive sampling near equilibrium.
The FE-NES implementation on the Orion® cloud platform automates these calculations, completing them within a few hours even for large ligand sets. This makes FE-NES ideal for iterative and large-scale lead optimization workflows.
With OpenEye’s automated solution, you gain speed, accuracy, and performance, while saving both time and cost.
"... 40 ligands could take 24-36 hours with FEP+ but only 2-3 hours with FE-NES on Orion" - Scientist, CADD, Large Pharma.
Features
- Accurate. Validated on internal and external datasets
- Fast. Extremely fast and highly parallelizable; run an entire ligand dataset in a few hours
- Scalable. Evaluate more analogs, faster, without compromising accuracy
- Flexible. Edge mapper options for connecting ligands include efficient star maps and networked OELOMAPs with cycle closure
- Easy-to-Use. Quickly get up and running in a familiar web-based environment
- Automated. Simple process from system set-up, mapping, end-point equilibration, and free energy calculation
- Control. Highly customizable for novice use and expert control
- Integrated. Easily combine with other AI and ligand- and structure-based methods
Rooted in Science, Optimized for Performance, Made for You
With a focus on cutting-edge science and technology, FE-NES simplifies the process of drug discovery by delivering powerful tools directly to your web browser on the Orion cloud modeling platform. There's no software to install; all you need is a web browser. Simply log in and get started!
Our automated solution is designed for speed, performance, and accuracy, helping you save both time and money.

Accurately Ranks Ligands Relative Binding Free Energies
FE-NES accurately ranks ligands by their relative binding free energies to accelerate lead optimization. Kendall’s tau correlations show that OpenEye's FE-NES method delivers accuracy comparable to other commercial and open-source approaches. Using the Schindler (2020) and Wang (2015) benchmark datasets from the literature, results demonstrate that FE-NES shows no significant differences in aggregate performance, making it a reliable solution for free energy calculations, with the added benefits of greater speed and lower computational cost.
Datasets: 1) Wang et al., 2015, J. Am. Chem. Soc., 137: 2695. and 2) Schindler et al., 2020, J. Chem. Inf. Model., 60: 5457
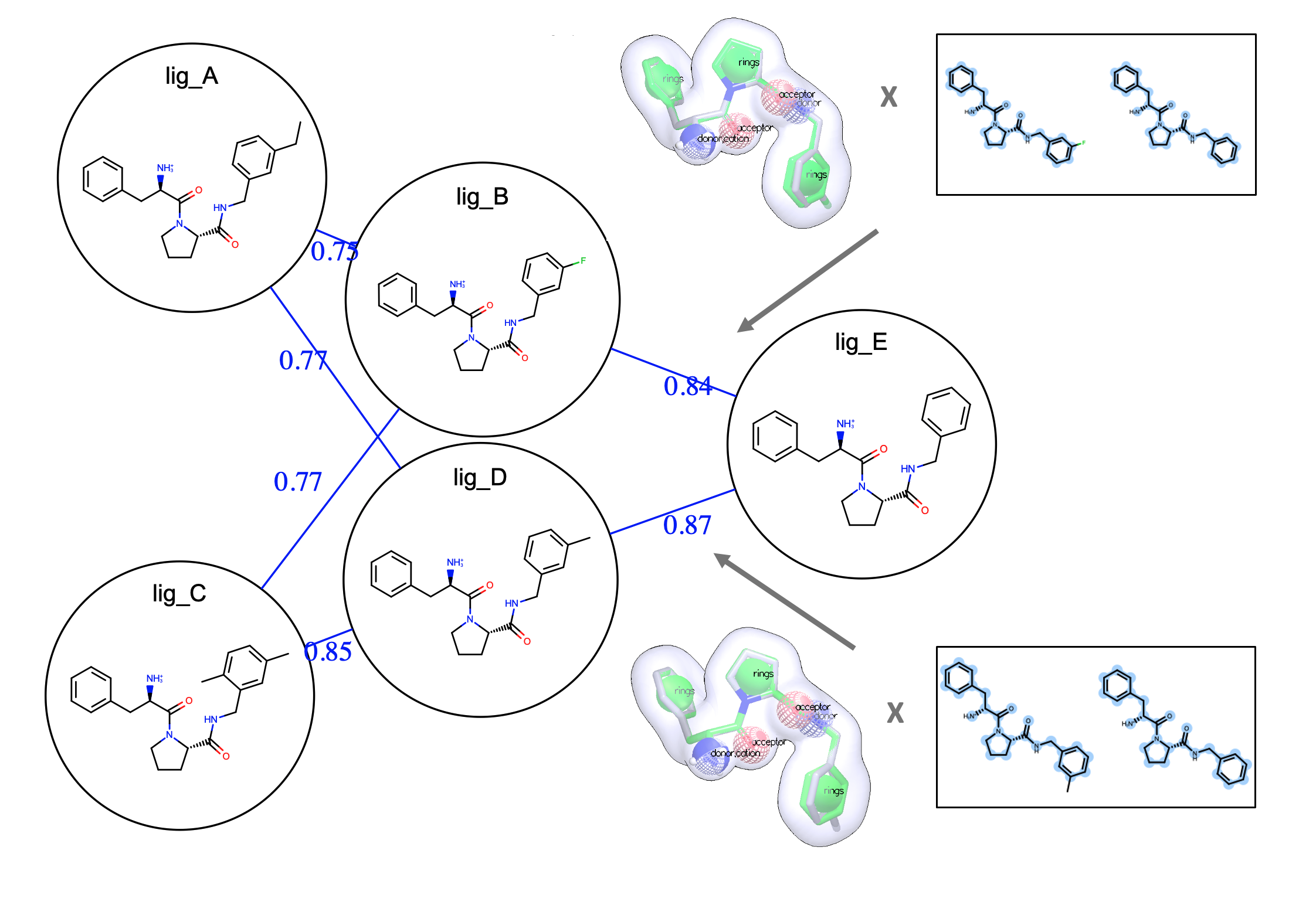
Easy, Efficient, and Automated Mapping
With FE-NES, users can easily create a map using a variety of mapper options including OELOMAP, Star Map, and Binary Star Map, and Multi-Star Map. The automated edge mappers connect ligands in binding free energy calculations.

Offering Flexibility with the Interactive Edge Mapper
Using the FE-NES interactive edge mapper, users can seamlessly add or remove edges and generate new maps with the selected ligand. The updated edge map can be saved as for downstream free energy calculations, allowing users to enhance the edge map with human expertise.

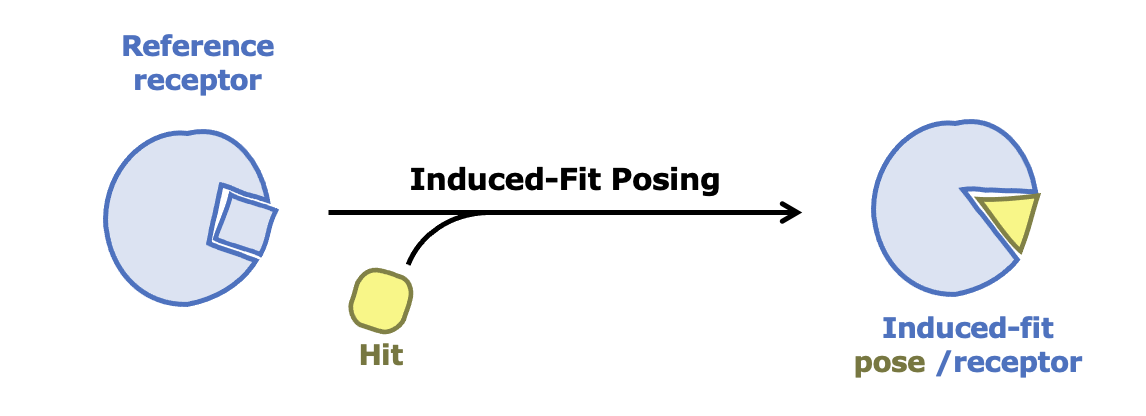
Improving Accuracy for Proteins with Flexible Binding Sites
Drug targets sometimes involve proteins with flexible binding sites, which can pose a challenge for accurate modeling. To improve predictions of ligand binding affinities in such cases, combine Induced-Fit Posing (IFP) with Free Energy Nonequilibrium Switching (FE-NES). Together, these tools deliver more accurate and reliable models of ligand–protein interactions.
Learn more about Induced-Fit Posing
FAQs
-
Yes, FE-NES supports alchemical changes between ligands of different formal charges.
-
FE-NES offers versatile workflows for a personalized experience, with options for both novice and expert users. Choose from pre-configured automated workflows or individual workflows for customized tasks with expert control.
-
Users get a comprehensive and interactive report of binding energy calculations. In addition, users can also seamlessly compare experimental affinity with the computed End Point Analysis results (predicted relative binding free energies).
-
FE-NES offers fast and affordable free energy calculations with its Nonequilibrium Switching approach.
Industry scientists have reported that FE-NES delivers market-leading accuracy while being 5–10X higher throughput and 2–5X more cost-effective than traditional equilibrium methods.
-
FE-NES, a highly efficient and cost-effective free energy calculation method, is adapted from the NES approach advanced by Vytautas Gapsys in the Bert L. DeGroot Lab at the Max Planck Institute for Biophysical Chemistry. (Gapsys et al., Chem. Sci. 2020, 11, 1140-1152). The method is highly parallelizeable making it suitable for high throughput calculations.
With the integration of proprietary OpenEye methods for system pre-equilibration and chimeric-molecule calculation, it delivers quick transitions through intermediate states and parallel sampling of short trajectories. This results in very fast, yet equally accurate calculations compared to conventional equilibrium-based methods.
-
OpenEye’s FE-NES method delivers accuracy comparable to other commercial and open-source approaches on public datasets such as Schindler (2020) and Wang (2015). On these datasets FE-NES shows no significant differences in aggregate performance to equilibrium methods such as FEP and TI, making it a reliable solution for free energy calculations, with the added benefits of greater speed and lower computational cost.
Datasets: 1) Wang et al., 2015, J. Am. Chem. Soc., 137: 2695. and 2) Schindler et al., 2020, J. Chem. Inf. Model., 60: 5457
-
Yes, FE-NES supports the use of the bespoke force field option.
-
FE-NES provides users with a Maximum Likelihood Estimator method to predict the absolute binding affinities starting from the relative binding affinities and a set of reference experimental binding free energies. This estimated value will be provided only if the graph edges are well connected and some experimental binding free energies are provided.
Learn More
FE-NES helps you rapidly and accurately estimate relative binding free energies, accelerating your drug design process and reducing costs by accurately ranking ligands to guide lead optimization.
For science details, access recording for OpenEye’s miniWebinar from November 12, 2025, on "Free energy prediction for drug discovery: Ideas at breakfast, discoveries by lunch" with Chris Neale, Ph.D.
Download the Science Brief to discover how Free Energy Nonequilibrium Switching (FE-NES) provides an accurate, fast and highly scalable method for calculating relative binding free energies.
References
-
Large scale relative protein ligand binding affinities using non-equilibrium alchemy, Gapsys et al., Chemical Science, 2020, 11, 1140-1152
-
Calculation of binding free energies, Gapsys et al., Methods Mol Biol., 2015, 1215:173-209
-
Accurate and reliable prediction of relative ligand binding potency in prospective drug discovery by way of a modern free-energy calculation protocol and force field. Wang et al., J Am Chem Soc. 2015 Feb 25;137(7):2695-703
-
Large-Scale Assessment of Binding Free Energy Calculations in Active Drug Discovery Projects. Schindler et al., J Chem Inf Model. 2020 Nov 23;60(11):5457-5474.
Webinar: OpenEye's Free energy prediction for drug discovery: Ideas at breakfast, discoveries by lunch
Science Brief: Binding Free Energy Redefined: Accurate, Fast, Affordable
CUP XXV - Santa Fe March 10-12, 2026
Webinar: Novel Hits from Beyond the Known: ROCS X
Resources
Glimpse the Future through News, Events, Webinars and more
News
ROCS X: AI-Enabled Molecular Search Unlocks Trillions
Webinar
Webinar: OpenEye's Free energy prediction for drug discovery: Ideas at breakfast, discoveries by lunch



