
SZYBKI/FreeForm
SZYBKI optimizes molecular structures with a force field, either with or without a solvent model, to yield quality 3D molecular structures for use as input to other programs. Good quality 3D conformations, either of a small molecule alone or in complex with a protein receptor, are critical for a number of key tasks including model building and shape and electrostatic comparisons.
Features
- The ability to perform ligand entropy calculations in different environments
- The ability to handle molecules containing selenium and boron.
- Faster calculation of default VdW protein-ligand potential in the active site via the use of lookup tables. Exact VdW protein-ligand potentials can still be calculated if desired
- Choice of force fields including OpenFF (Open Force Field Initiative) force fields Sage and Parsley, AMBER Protein force fields ff14SB and AMBER99, and the MMFF94 (Merck Molecular Force Field)[2]
- Solvent effects with the high accuracy Poisson-Boltzmann model or the faster Sheffield model for binding energies and geometry optimization
- Flexibility of protein active-site polar hydrogens or side-chains
- Ligand optimization within the field of a protein active site
- Minimization in several coordinate frames (Cartesian, torsional, rigid body)
- Fully optimizes (gas phase) about 200 drug-sized molecules per minute
- Optionally excludes various terms from potential when determining energy and gradient
- Constrains coordinates (fixed atoms or harmonic constraints)
- Distributed processing via MPI for all supported platforms
![Accurate Poisson—Boltzmann vs. Sheffield Solvation Model solvation energies for a database of 64,000 drug-like molecules [3].](https://www.eyesopen.com/hs-fs/hubfs/szybki-graph.jpg?width=623&height=462&name=szybki-graph.jpg)
SZYBKI also refines portions of a protein structure and optimizes ligands within a protein active site, making it useful in conjunction with docking programs.
Supported force fields in SZYBKI include OpenFF (Open Force Field Initiative) force fields Sage and Parsley, AMBER Protein force fields ff14SB and AMBER99, and the MMFF94 (Merck Molecular Force Field).
FreeForm is a utility program included with SZYBKI that provides two functionalities:
- calculates the solvation energy of the free ligand, and
- calculates the free energy of going from an ensemble of solution phase conformers to a single, bioactive conformation.
For more detailed information on SZYBKI, check out the link below:
Documentation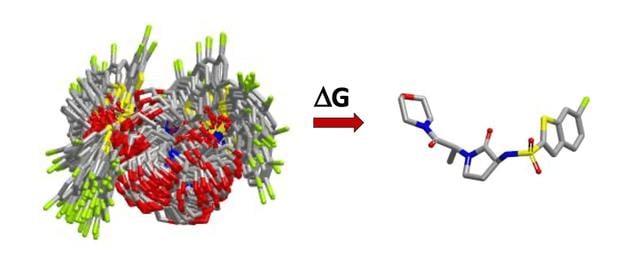
References
- Szybki (shib´·kee) means "fast" in Polish.
- Ligand entropy in gas-phase, upon solvation and protein complexation. Fast estimation with Quasi-Newton Hessian Wlodek, S., Skillman, A.G., Nicholls, A. J. Chem. Theory Comput., 2010, 6(7), 2140-2152.
- Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94 Halgren, T.A., J. Comp. Chem, 1996, 17, 490. MMFF94s option for energy minimization studies Halgren, T.A., J. Comp. Chem, 1999, 20, 720.
- A simple formula for dielectric polarisation energies: The Sheffield Solvation Model Grant, J.A., Pickup, B.T., Sykes, M.J., Kitchen, C.A., Nicholls, A., Chem. Phys. Lett., 2007, 441, 163.
RESOURCES
