
OEChem TK
OEChem TK is a programming library for chemistry and cheminformatics that is fast and flexible. OEChem TK has many simple yet powerful functions that handle the details of working with small molecules, as well as an expanding number of functions for dealing with proteins. High-level functions provide simplicity while low-level functions provide flexibility.
The OEChem TK also includes two sub-libraries designed to handle macromolecules (OEBio) and grids (OEGrid).
After several failed attempts with open source software, I resorted to a commercial solution. The best tool I found for parsing XYZ files and assigning bond orders was the OEChem toolkit from OpenEye Scientific Software. Using OEChem, I could parse the XYZ files and correctly assign bond orders. - Pat Walters, Practical Cheminformatics
Features
- Facile management of molecules, atoms, bonds, and conformers
- Conformational and frame-of-reference coordinate transformations
- Maximum common substructure
- Substructure searching based on SMARTS or MDL query
- Perception of aromaticity with multiple models
- Chemical reaction parsing and processing
- Library generation based on SMIRKS or MDL reaction
- Tetrahedral and E/Z stereochemistry recognition
- CIP atom and bond stereo perception
- Ring perception and Kekulization
- Molecular canonicalization
- Multiconformer molecule handling
- Ability to store and recall generic primitives or user-defined objects on molecules, atoms, bonds, or conformers
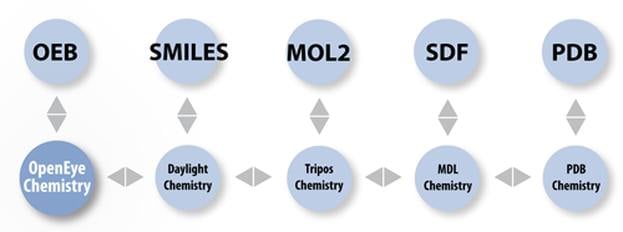
| File Format | read | write |
| OpenEye's binary | Yes | Yes |
| MDL Mol | Yes | Yes |
| MDL SD | Yes | Yes |
| MDL RDF | Yes | No |
| Protein Databank PDB | Yes | Yes |
| Tripos Sybyl mol2 | Yes | Yes |
| Canonical SMILES | Yes | Yes |
| Canonical isomeric SMILES | Yes | Yes |
| InChi | No | Yes |
| InChiKey | No | Yes |
| FASTA protein sequence | Yes | Yes |
| Macromodel | Yes | Yes |
| XMol XYZ | Yes | Yes |
| CIF/mmCIF | Yes | Yes |
Key Features of OEBio:
- Protein residue, the primary, secondary and tertiary structure hierarchy perception
- Crystal symmetry handling
- Sequence alignment
- Management of torsions, rotamer libraries, and alternate conformations
Key Features of OEGrid:
- Support for the following grid file formats: Grasp, GRD (OpenEye Binary format), CCP4, XPLOR
For more detailed information on OEChem TK, check out the link below:
Modeling
The Modeling suite of toolkits provides the core functionality underlying OpenEye's defining principle that shape & electrostatics are the two fundamental descriptors determining intermolecular interactions. Many of the toolkits in the Modeling suite are directly associated with specific OpenEye applications and can, therefore, be used to create new or extend existing functionality associated with those applications.
- OEChem TK Core chemistry handling and representation as well as molecule file I/O
- FastROCS™ TK Real-time shape similarity for virtual screening, lead hopping & shape clustering
- OEDocking TK Molecular docking and scoring
- Omega TK Conformer generation
- Shape TK 3D shape description, optimization, and overlap
- SiteHopper TK Rapid comparison of protein binding sites
- Spicoli TK Surface generation, manipulation, and interrogation
- Spruce TK Protein preparation and modeling
- Szybki TK Force field based focused optimization and entropy estimations
- OEFF TK Force fields and general optimization tools
- Szmap TK Understanding water interactions in a binding site
- Zap TK Calculate Poisson-Boltzmann electrostatic potentials
- Bioisostere TK 3D fragment similarity, lead optimization, and analog generation
- Eon TK Molecular shape and electrostatic overlap
Cheminformatics
The Cheminformatics suite of toolkits provides the core foundation upon which all the OpenEye applications and remaining toolkits are built.
- OEChem TK Core chemistry handling and representation as well as molecule file I/O
- OEDepict TK 2D Molecule rendering and depiction
- Grapheme™ TK Advanced molecule rendering and report generation
- GraphSim TK 2D molecular similarity (e.g. fingerprints)
- Lexichem™ TK Name-to-structure, structure-to-name, foreign language translation
- MolProp TK Molecular property calculation and filtering
- Quacpac TK Tautomer enumeration and charge assignment
- OEMedChem TK Matched molecular pair analysis, fragmentation utilities, and molecular complexity metrics
- Saiph TK: Extracting, transforming, and loading (ETL) files and records
References
- Optimizing Fragment and Scaffold Docking by Use of Molecular Interaction Fingerprints Gilles Marcou, Didier Rognan. J. Chem. Inf. Model., 2007, 47 (1), 195-207.
- Database Clustering with a Combination of Fingerprint and Maximum Common Substructure Methods Martin Stahl, Harald Mauser. J. Chem. Inf. Model., 2005, 45 (3), 542-548.
Webinar: OpenEye's Free energy prediction for drug discovery: Ideas at breakfast, discoveries by lunch
Science Brief: Binding Free Energy Redefined: Accurate, Fast, Affordable
CUP XXV - Santa Fe March 10-12, 2026
Webinar: Novel Hits from Beyond the Known: ROCS X
Resources
Glimpse the Future through News, Events, Webinars and more
News
ROCS X: AI-Enabled Molecular Search Unlocks Trillions
Webinar
Webinar: OpenEye's Free energy prediction for drug discovery: Ideas at breakfast, discoveries by lunch


