
CALCULATING MOLECULAR SIMILARITY
GraphSim TK
Measuring molecular similarity and diversity plays an important role in various steps of the drug design cycle. Calculating molecule similarity is extensively used in applications such as virtual screening, property prediction, synthesis design and chemical database clustering.
Fingerprinting provides an elementary encoding of molecular graphs. Even though fingerprints can only represent local structural features and not their relative positions in molecules, it has proven to be very successful in a range of similarity and diversity studies.
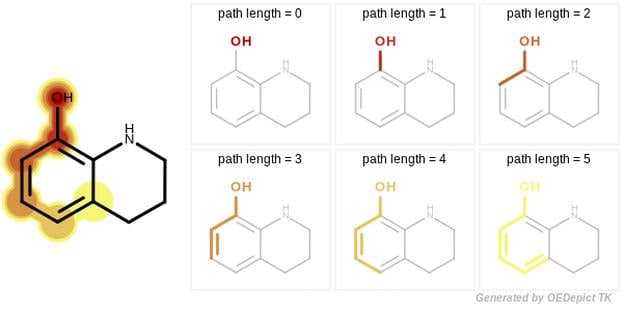
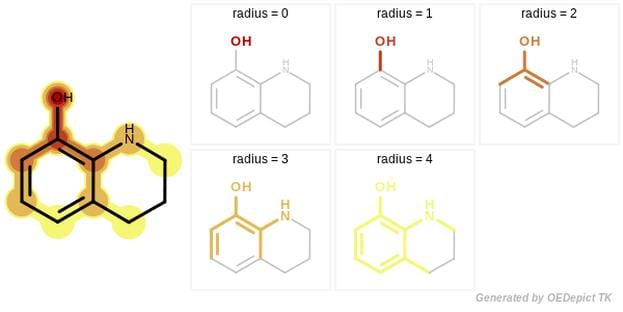
GraphSim TK provides five different fingerprint types to perform 2D molecular similarity measurements:
- Path
- Circular [1]
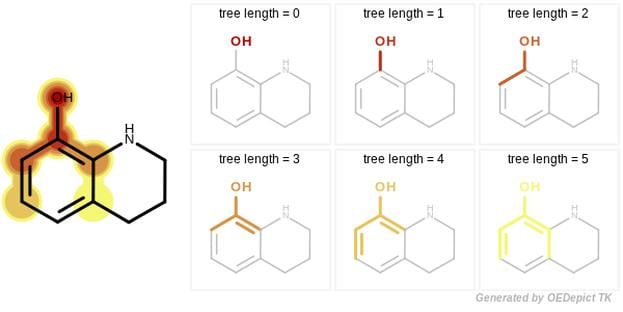
- Tree
- MACCS key [2]
- LINGO [3]
A path fingerprint is generated by exhaustively enumerating all linear fragments of a molecular graph up to a given size and hashing these fragments into a fixed-length bivector.
The tree fingerprint provides a novel approach that encodes molecular motifs that can not be captured by the Path or the Circular methods. This involves exhaustively enumerating all unique trees of a molecular graph.
The novelty of the GraphSim TK fingerprint generation is that it gives full control to the user to specify the size of the enumerated paths, along with the atom and bond properties that are used when path, circular or tree fragments are encoded into bitvectors.
The fingerprint methods were parameterized and validated using two published benchmarks:
- Briem-Lessel [4] (5 activity classes _ small set of decoys)
- Hert- Willet [5] (11 activity classes + entire MDDR used as a set of inactive compounds)
Measuring molecular similarity or dissimilarity has two basic components:
- the representation of molecular characteristics (such as fingerprints) and
- the similarity coefficient that is used to quantify the degree of resemblance between two such representations.
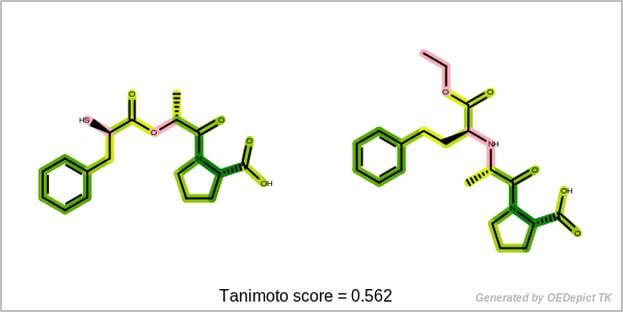
GraphSim TK supports several built-in similarity coefficients (Cosine, Dice, Euclidean, Manhattan, Tanimoto, Tversky), while user-defined similarity measures are also available.
Apart from generating and storing fingerprints, GraphSim TK also provides a fingerprint database that is designed to perform rapid in-memory fingerprint search utilizing any of the built-in or user-defined similarity measures.
GraphSim TK also provides access to the common fragments found between two molecules based on the given fingerprint type.
This data can be used to visualize the similarity of two molecules based on their fingerprints. The fingerprint overlap, depicted in 2D, provides insight into molecule similarity beyond a single numerical score and reveals information about the underlying fingerprint method.
For more detailed information on GraphSim TK, check out the link below:
DocumentationCheminformatics
The Cheminformatics suite of toolkits provides the core foundation upon which all of the OpenEye applications and remaining toolkits are built. The Cheminformatics suite is a collection of seven individual yet interdependent toolkits that are described in the table below.
- FastROCS TK Real-time shape similarity for virtual screening, lead hopping & shape clustering
- OEChem TK Core chemistry handling and representation as well as molecule file I/O
- OEDepict TK 2D Molecule rendering and depiction
- Grapheme™ TK Advanced molecule rendering and report generation
- GraphSim TK 2D molecular similarity (e.g. fingerprints)
- Lexichem TK name-to-structure, structure-to-name, foreign language translation
- MolProp TK Molecular property calculation and filtering
- Quacpac TK Tautomer enumeration and charge assignment
- MedChem TK Matched molecular pair analysis, fragmentation utilities, and molecular complexity metrics
Modeling
The Modeling suite of toolkits provides the core functionality underlying OpenEye's defining principle that shape & electrostatics are the two fundamental descriptors determining intermolecular interactions. Many of the toolkits in the Modeling suite are directly associated with specific OpenEye applications and can therefore be used to create new or extend existing functionality associated with those applications.
- OEChem TK Core chemistry handling and representation as well as molecule file I/O
- OEDocking TK Molecular docking and scoring
- Omega TK Conformer generation
- Shape TK 3D shape description, optimization, and overlap
- SiteHopper TK Rapid Comparison of Protein Binding Sites
- Spicoli TK Surface generation, manipulation, and interrogation
- Spruce TK Protein preparation and modeling
- Szybki TK General purpose optimization with MMFF94
- Szmap TK Understanding water interactions in a binding site
- Zap TK Calculate Poisson-Boltzmann electrostatic potentials
References
- Extended-Connectivity Fingerprints, D. Rogers, M. Hahn. J. Chem. Inf. Model., 2010, 50, (5) 742-754.
- Reoptimization of MDL Keys for Use in Drug Discovery, J. L. Durant, B. A. Leland, D. R. Henry, J. G. Nourse. J. Chem. Inf. Comput. Sci., 2002, 42, (6) 1273-1280.
- LINGO, an Efficient Holographic Text Based Method To Calculate Biophysical Properties and Intermolecular Similarities, D. Vidal, M. Thormann, M. Pons. J. Chem. Inf. Model., 2005,45, (2) 386-393.
- In vitro and in silico affinity fingerprints: Finding similarities beyond structural classes, H. Briem, U. F. Lessel. Perspectives in Drug Discovery and Design, 2004, 20, 231-244.
- Comparison of Fingerprint-Based Methods for Virtual Screening Using Multiple Bioactive Reference Structures, J. Hert, P. Willett, D. J. Wilton. J. Chem. Inf. Comput. Sci., 2004, 44, (3) 1177-1185.
RESOURCES
