
Modeling Development Platform
The Modeling suite of toolkits provides the core functionality underlying OpenEye's defining principle that shape & electrostatics are the two fundamental descriptors determining intermolecular interactions.
Many of the toolkits in the Modeling suite are directly associated with specific OpenEye applications and can, therefore be used to create new or extend existing functionality associated with those applications.
Sitehopper, a shape-based, binding site comparison tool is an excellent example of using the Modeling Toolkits to create new methods.
For more detailed information on the Modeling Development Platform, check out the link below:
Documentation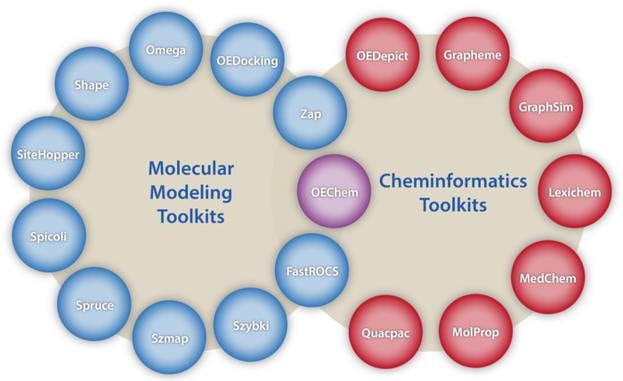
Modeling
The Modeling suite of toolkits provides the core functionality underlying OpenEye's defining principle that shape & electrostatics are the two fundamental descriptors determining intermolecular interactions. Many of the toolkits in the Modeling suite are directly associated with specific OpenEye applications and can, therefore, be used to create new or extend existing functionality associated with those applications.
- OEChem TK Core chemistry handling and representation as well as molecule file I/O
- FastROCS™ TK Real-time shape similarity for virtual screening, lead hopping & shape clustering
- OEDocking TK Molecular docking and scoring
- Omega TK Conformer generation
- Shape TK 3D shape description, optimization, and overlap
- SiteHopper TK Rapid Comparison of Protein Binding Sites
- Spicoli TK Surface generation, manipulation, and interrogation
- Spruce TK Protein preparation and modeling
- Szybki TK General purpose optimization with MMFF94
- Szmap TK Understanding water interactions in a binding site
- Zap TK Calculate Poisson-Boltzmann electrostatic potentials
Cheminformatics
The Cheminformatics suite of toolkits provides the core foundation upon which all the OpenEye applications and remaining toolkits are built.
- OEChem TK Core chemistry handling and representation as well as molecule file I/O
- OEDepict TK 2D Molecule rendering and depiction
- Grapheme™ TK Advanced molecule rendering and report generation
- GraphSim TK 2D molecular similarity (e.g. fingerprints)
- Lexichem TK name-to-structure, structure-to-name, foreign language translation
- MolProp TK Molecular property calculation and filtering
- Quacpac TK Tautomer enumeration and charge assignment
- MedChem TK Matched molecular pair analysis, fragmentation utilities, and molecular complexity metrics
Getting Started
In addition to the extensive API documentation provided for each toolkit, we also provide a Python Cookbook containing an ever growing collection of solutions and practical examples for cheminformatics and molecular modeling problems.
To learn more about what others are doing, please visit our Integration Partners page showcasing a wide variety of organizations using OpenEye tools to provide their own custom solutions and services.
Webinar: OpenEye's Free energy prediction for drug discovery: Ideas at breakfast, discoveries by lunch
Science Brief: Binding Free Energy Redefined: Accurate, Fast, Affordable
CUP XXV - Santa Fe March 10-12, 2026
Webinar: Novel Hits from Beyond the Known: ROCS X
Resources
Glimpse the Future through News, Events, Webinars and more
News
ROCS X: AI-Enabled Molecular Search Unlocks Trillions
Webinar
Webinar: OpenEye's Free energy prediction for drug discovery: Ideas at breakfast, discoveries by lunch


