OpenEye is pleased to announce the release of OMEGA v3.0.0. This major release adds many significant new features and fixes several bugs.
HIGHLIGHTS
- OMEGA’s capability has been expanded for molecules containing large rings by adding a method specifically tuned to sample macrocyclic conformational space. The approach is based on a rewritten version of the original OMEGA distance geometry algorithm.
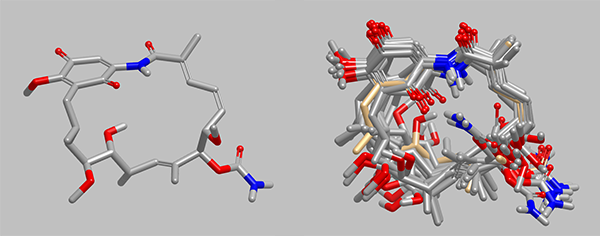
Conformer ensemble for geldanamycin, an inhibitor of heat shock protein 90, Hsp90 (PDB: 1YET),
generated using the new macrocycle method
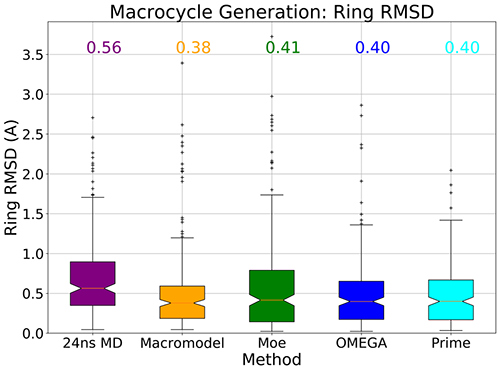
Median RMSD for reproduction of ring atom geometry in a set of 205 solid-state structures of macrocycles
- OMEGA now includes five modes:
- classic: the original customizable OMEGA interface
- macrocycle: the conformer generator for macrocycles
- rocs: the optimal conformer generator for ROCS
- pose: the optimal conformer generator for pose prediction or docking
- dense: the optimal conformer generator for FREEFORM
- A new torsion library has been added to OMEGA. This library is based on a 2016 probabilistic analysis of torsion angles in the CSD by Guba and co-workers. Introduced in the 2017.Oct Toolkits Release, this torsion library contains more than 500 new entries and complements the existing library, which is primarily based on an analysis of structures found in the PDB. A new flag, -torlibtype, has been added to the OMEGA classic mode to allow users to specify which of the two libraries should be used.
- New options have been added to the –buildff and -searchff flags to modify the MMFF94S force field to produce equatorial conformers for mono-substituted cyclohexanes when these are required but not normally produced.
GENERAL
- OMEGA and the ancillary programs are built on top of the OpenEye Toolkits v2017.Oct libraries to ensure that all the programs are taking advantage of state-of-the-art improvements in the underlying programming libraries.
PLATFORM SUPPORT
- Support for 64-bit Windows has been added.
- 32-bit Windows is no longer supported.
- Support for Ubuntu 14 and 16 has been added.
- Ubuntu 12 is no longer supported.
- Support for RedHat v7 has been added.
- RedHat v5 is no longer supported.
- Support for macOS El Capitan (10.11), macOS Sierra (10.12), and macOS High Sierra (10.13) has been added.
AVAILABILITY
OMEGA is now available for download. Existing licenses will continue to work, but if a new license is required, please contact your account manager or email sales@eyesopen.com.
See the release notes for full and specific details on improvements and fixes.
RESOURCES
About OpenEye Scientific
OpenEye Scientific is a privately held company headquartered in Santa Fe, New Mexico, with offices in Boston, Cologne, and Tokyo. It was founded in 1997 to develop large-scale molecular modeling applications and toolkits. Primarily aimed towards drug discovery and design, areas of application include:
- Cheminformatics
- Structure Generation
- Shape Comparison
- Docking
- Fragment Replacement
- Electrostatics
- Crystallography
- Visualization
The software is designed for scientific rigor as well as speed, scalability, and platform independence. OpenEye makes most of its technology available as toolkits—programming libraries suitable for custom development. OpenEye software typically is distributable across multiple processors and runs on Linux, Windows, and macOS.
For additional information
Jeffrey Grandy
Vice President, Sales
+1-415-863-3032
Email: sales@eyesopen.com

