
OEDocking TK
There are a wide variety of docking programs available to the community at large; however, until now there have been no resources in existence for individual methods developers to provide a common and well-supported foundation for the development of new docking and scoring applications.
The OEDocking TK from OpenEye is a programming library that provides this core docking and scoring functionality. Coupled with the highly acclaimed cheminformatics functionality in OEChem, the OEDocking TK is a comprehensive solution for anyone seeking to develop new docking tools. The toolkit supports docking, scoring, and optimization with the Chemscore, Chemgauss3, PLP, and Shapegauss scoring functions. The ligand-aware Hybrid Docking functionality unique to OpenEye's FRED docking program as well as the POSIT shape fitting algorithm for pose prediction are also provided in the toolkit. C++, C#, Python, and Java are supported.
For more detailed information on OEDocking TK, check out the link below:
Documentation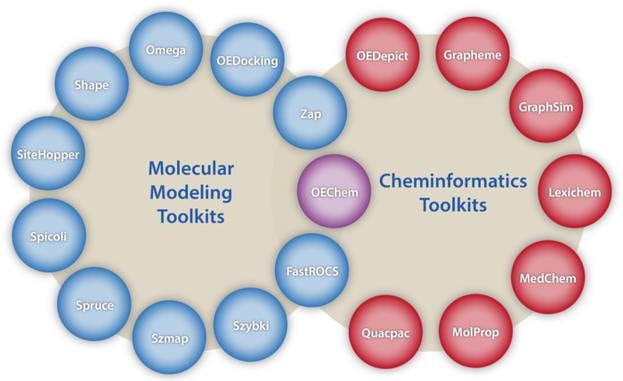
Features
Docking
- Exhaustive search followed by pose optimization
- Hybrid docking (uses the structure of a known bound active to guide docking)
- Docking constraints
Scoring
- Score optimization (systematic solid body optimization)
- Breakdown of score by atom and/or scoring function component
- Score annotation (scores are stored on molecule for visualization in VIDA
Scoring Functions
- Chemgauss4
- Chemgauss3
- Chemscore
- PLP
- Shapegauss
Modeling
The Modeling suite of toolkits provides the core functionality underlying OpenEye's defining principle that shape & electrostatics are the two fundamental descriptors determining intermolecular interactions. Many of the toolkits in the Modeling suite are directly associated with specific OpenEye applications and can, therefore, be used to create new or extend existing functionality associated with those applications.
- OEChem TK Core chemistry handling and representation as well as molecule file I/O
- FastROCS™ TK Real-time shape similarity for virtual screening, lead hopping & shape clustering
- OEDocking TK Molecular docking and scoring
- Omega TK Conformer generation
- Shape TK 3D shape description, optimization, and overlap
- SiteHopper TK Rapid comparison of protein binding sites
- Spicoli TK Surface generation, manipulation, and interrogation
- Spruce TK Protein preparation and modeling
- Szybki TK Force field based focused optimization and entropy estimations
- OEFF TK Force fields and general optimization tools
- Szmap TK Understanding water interactions in a binding site
- Zap TK Calculate Poisson-Boltzmann electrostatic potentials
- Bioisostere TK 3D fragment similarity, lead optimization, and analog generation
- Eon TK Molecular shape and electrostatic overlap
Cheminformatics
The Cheminformatics suite of toolkits provides the core foundation upon which all the OpenEye applications and remaining toolkits are built.
- OEChem TK Core chemistry handling and representation as well as molecule file I/O
- OEDepict TK 2D Molecule rendering and depiction
- Grapheme™ TK Advanced molecule rendering and report generation
- GraphSim TK 2D molecular similarity (e.g. fingerprints)
- Lexichem™ TK Name-to-structure, structure-to-name, foreign language translation
- MolProp TK Molecular property calculation and filtering
- Quacpac TK Tautomer enumeration and charge assignment
- OEMedChem TK Matched molecular pair analysis, fragmentation utilities, and molecular complexity metrics
- Saiph TK: Extracting, transforming, and loading (ETL) files and records
References
- Gaussian Docking Functions Mark McGann, Harold R Almond, Anthony Nicholls, J. Andrew Grant and Frank K. Brown, BioPolymers, 2003, 68, 76-90.
- Deciphering common failures in molecular docking of ligand-protein complexes Gennady M. Verkivker, Djamal Bouzida, Daniel K. Gehlaar, Paul A. Rejto, Sandra Arthurs, Anthony B. Colson, Stephan T. Freer, Veda Larson, Brock A. Luty, Tami Marrone and Peter W. Rose, J. Comput. Aided Mol. Des., 2000, 14, 731-751.
- Empirical scoring functions: I. The development of a fast empirical scoring function to estimate the binding affinity of ligands in receptor complexes Matthew D. Eldridge, Christopher W. Murray, Timothy R. Auton, Gaia V. Paolini and Roger P. Mee., J. Comput. Aided Mol. Des., 1997, 11, 425-445.
Webinar: OpenEye's Free energy prediction for drug discovery: Ideas at breakfast, discoveries by lunch
Science Brief: Binding Free Energy Redefined: Accurate, Fast, Affordable
CUP XXV - Santa Fe March 10-12, 2026
Webinar: Novel Hits from Beyond the Known: ROCS X
Resources
Glimpse the Future through News, Events, Webinars and more
News
ROCS X: AI-Enabled Molecular Search Unlocks Trillions
Webinar
Webinar: OpenEye's Free energy prediction for drug discovery: Ideas at breakfast, discoveries by lunch


