OpenEye is pleased to announce the release of OpenEye Toolkits v2017.Oct. These libraries include the usual support for C++, Python, C#, and Java.
EXAMPLE NEW FEATURES
- The 2017.Oct release introduces Shape TK version 2.0, a complete redesign of Shape TK.
- A new torsion library has been added to Omega TK.
- OEDepict TK and Grapheme TK now have expanded capability to generate interactive images that are both simple and information-rich.
Shape TK version 2.0
The 2017.Oct release introduces Shape TK version 2.0. This new version has been completely redesigned to make the library thread-safe, more flexible, and allows the new API to be easily extended with additional functionality. Overlay optimization in Shape TK version 2.0 is ~10% faster than the 2017.Jun version.
New features in Shape TK version 2.0 include the ability to:
- calculate color using grid- or proxy grid-based analytic methods
- calculate shape overlap and color score between two fixed-in-space objects with a single calculation using the OEOverlapFunc class
- combine force fields from OEFF TK with shape and/or color for overlay optimization
- obtain top hits from a database against a given query using the OEROCS class
- create a query using any combination of a molecule, a grid, and multiple shape and/or color Gaussians using the OEShapeQuery class (see image below)
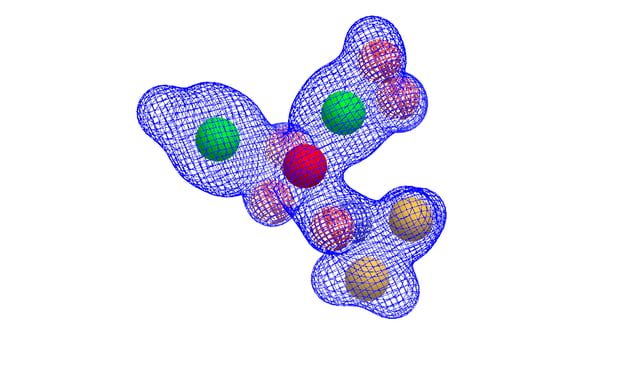
Shape query with shape grid and color Gaussians
Omega TK
A new torsion library has been added to Omega TK based on a 2016 probabilistic analysis of torsion angles in the Cambridge Structural Database (CSD) by Guba and co-workers. This torsion library, which contains more than 500 new entries, complements the existing library, which is based mainly on an analysis of structures found in the Protein Data Bank (PDB). Replacing the existing library has four important effects on Omega’s performance:
- near elimination of torsion values not well represented in the CSD
- significantly improved run time
- substantial reduction in the size of the conformer ensemble
- a small trade-off increase in the median RMSD for solid state structures (both from the PDB and the CSD)
OEDepict TK and Grapheme TK
The 2017.Oct release expands these toolkits’ capability to group a set of graphical objects in SVG images. First introduced in the 2017.Jun release, this feature can now generate interactive images. Users can glean embedded information by simply hovering a mouse over the desired area of the image.
For example, the sequence-level depiction of a peptide, cyclic trypsin inhibitor (PDB: 1JBL), is attractive due to its simplicity (see image below). The more detailed atomic-level representation of the amino acid components of the peptide can be revealed on mouse-over without cluttering the simplified view.
Peptide depiction of cyclic trypsin inhibitor (PDB: 1JBL)
PLATFORM SUPPORT
- On macOS, Python single-build distributions are now installed by default when using the meta-installer via pip install openeye-toolkits.
- The Python verification tests now use pytest rather than nosetest as the test discovery tool. pytest options can be passed to the openeye.examples.openeye_tests module to control the tests and level of debugging information provided.
- The 2017.Oct release is the last release to support Java 1.6.
- The 2017.Oct release is the last release to support Python 2.7.
Note: As this is the last release to support Python 2.7, OpenEye is willing to help with code migration. Please contact support@eyesopen.com for more details.
AVAILABILITY
OpenEye Toolkits v2017.Oct are now available for download. Existing licenses will continue to work, but if a new license is required, please contact your account manager or email sales@eyesopen.com.
See the Release Notes for full and specific details on improvements and fixes.
|
About OpenEye Scientific Software
OpenEye Scientific Software Inc. is a privately held company headquartered in Santa Fe, NM, with offices in Boston, Cologne, and Tokyo. It was founded in 1997 to develop large-scale molecular modeling applications and toolkits. Primarily aimed towards drug discovery and design, areas of application include:
- Cheminformatics
- Structure Generation
- Shape Comparison
- Docking
- Fragment Replacement
- Electrostatics
- Crystallography
- Visualization
The software is designed for scientific rigor, as well as speed, scalability and platform independence. OpenEye makes most of its technology available as toolkits - programming libraries suitable for custom development. OpenEye software typically is distributable across multiple processors and runs on Linux, Windows and Mac OS X.
For additional information
Jeffrey Grandy
VP Sales
+1-415-863-3032
Email: sales@eyesopen.com