OpenEye is pleased to announce the release of OpenEye Toolkits v2018.Feb. These libraries include the usual support for C++, Python, C#, and Java.
HIGHLIGHTS
- GraphSim TK now supports GPU-enabled fingerprint searching.
- Grapheme TK now supports unpaired interaction maps.
- Conda toolkit packages for Python are now fully supported.
GraphSim TK: GPU-Accelerated Fast Fingerprint Similarity Searches
CUDA support for faster fingerprint searching is now available in GraphSim TK. This functionality allows users to perform molecular similarity calculations up to 200x faster than the in-memory fast fingerprint mode (see the NxN similarity score calculation graph below). The CUDA mode involves preloading all fingerprints into GPU memory prior to performing similarity calculations. Searches are limited by GPU memory availability and will fall back to the memory-mapped CPU mode if the entire set of fingerprints cannot be preloaded into the GPU memory. Despite this limitation, this represents the fastest way to perform similarity searches.
The new OEFastFPDatabase::GetHistogram method performs an NxN search of a molecular database and returns a histogram of scores. This is useful information for analyzing collection diversity without needing disk space for NxN similarities. OEFastFPDatabase::GetHistogram in CUDA mode performs 2.5 billion comparisons per second (as benchmarked on an AWS P3 instance with an NVIDIA Tesla V100 GPU) compared with 12.5 million comparisons per second when using the memory-mapped mode on the CPU.
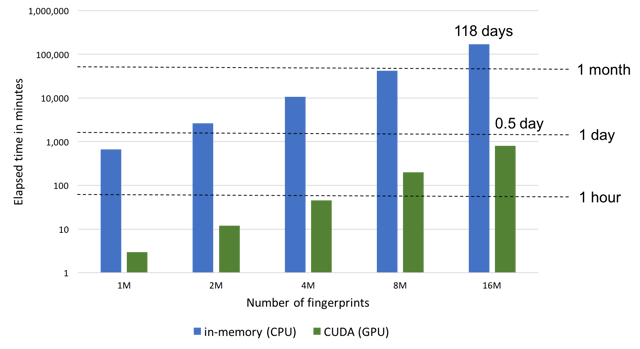
Performance of an NxN similarity score calculation. 16M 4096-bit long tree fingerprints = 8GB data. CPU benchmarked on m4.10xlarge AWS instance (single threaded). GPU benchmarked on p3.8xlarge AWS instance
![Performance-SimilaritySearch-CPU-GPU[3].png](https://www.eyesopen.com/hs-fs/hubfs/News-Events/Performance-SimilaritySearch-CPU-GPU%5B3%5D.png?width=640&name=Performance-SimilaritySearch-CPU-GPU%5B3%5D.png)
Performance of a fingerprint search (load time not included). 28M 4096-bit long tree fingerprints = 64GB data. Retrieving the 10 most similar molecules. CPU benchmarked on m4.10xlarge AWS instance (single threaded). GPU benchmarked on p3.8xlarge AWS instance
Grapheme TK: Unpaired Interaction Map
Grapheme TK now includes the ability to generate unpaired interaction maps. An interaction map (see the figure on the right below) depicts interactions currently made, while the unpaired map (see the figure on the left below) illustrates interactions that are not now formed but could potentially contribute to ligand binding. Together, these two maps of the protein-ligand binding site provide insights into ligand binding and communicate complex 3D structural results.
In an unpaired interaction map, different linker types are used to mark unpaired acceptor and donor hydrogen bond interactions. Since these interactions are unpaired, the direction of the linkers has no real spatial meaning. Ligand linkers are directed away from the ligand, while protein linkers are directed towards the ligand. The geometric parameters for unpaired interactions are customizable to fit user preferences.
Unpaired interaction map (left) and active site depiction (right) generated after hydrogen placement (PDB: 1BR6)
Conda Packages for Python
Conda toolkits for Python are now available as a package named openeye-toolkits in the openeye channel on the default conda.anaconda.org server. On Linux and macOS, the toolkits can be installed into a Conda Python environment via conda install -c openeye openeye-toolkits. On Windows, Conda packages can be managed using Anaconda Navigator.
GENERAL NOTICES
- All OpenEye Toolkit Python code examples now follow the PEP8
PLATFORM SUPPORT
- The 2018.Feb release fully supports macOS High Sierra 10.13.
- macOS 10.10 is no longer supported.
AVAILABILITY
OpenEye Toolkits v2018.Feb are now available for download. Existing licenses will continue to work, but if a new license is required, please contact your account manager or email sales@eyesopen.com.
See the Release Notes for full and specific details on improvements and fixes.
RESOURCES
|
About OpenEye Scientific
OpenEye Scientific is a privately held company headquartered in Santa Fe, New Mexico, with offices in Boston, Cologne, and Tokyo. It was founded in 1997 to develop large-scale molecular modeling applications and toolkits. Primarily aimed towards drug discovery and design, areas of application include:
- Cheminformatics
- Structure Generation
- Shape Comparison
- Docking
- Fragment Replacement
- Electrostatics
- Crystallography
- Visualization
The software is designed for scientific rigor as well as speed, scalability, and platform independence. OpenEye makes most of its technology available as toolkits—programming libraries suitable for custom development. OpenEye software typically is distributable across multiple processors and runs on Linux, Windows, and macOS.
For additional information
Jeffrey Grandy
VP Sales
+1-415-863-3032
Email: sales@eyesopen.com